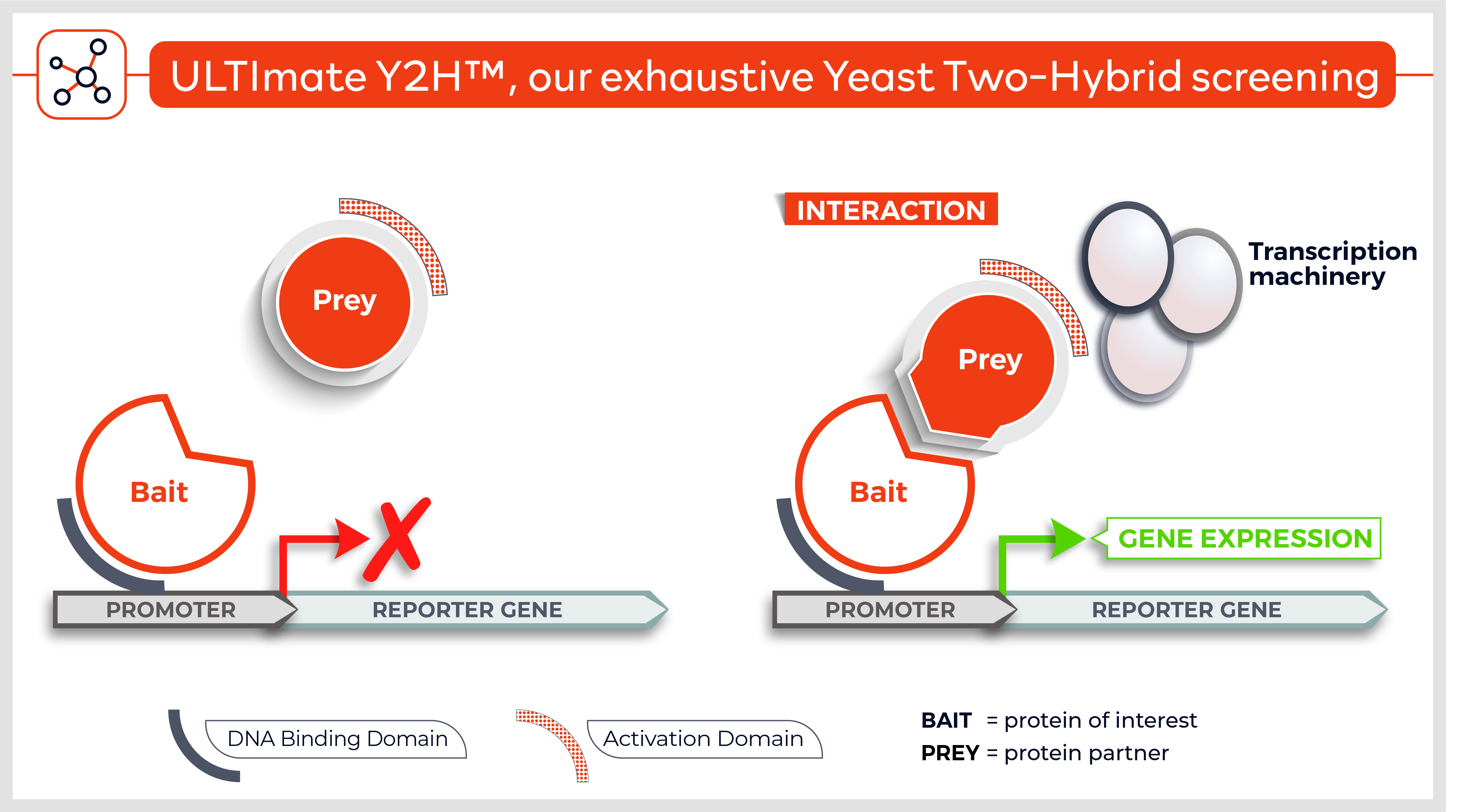
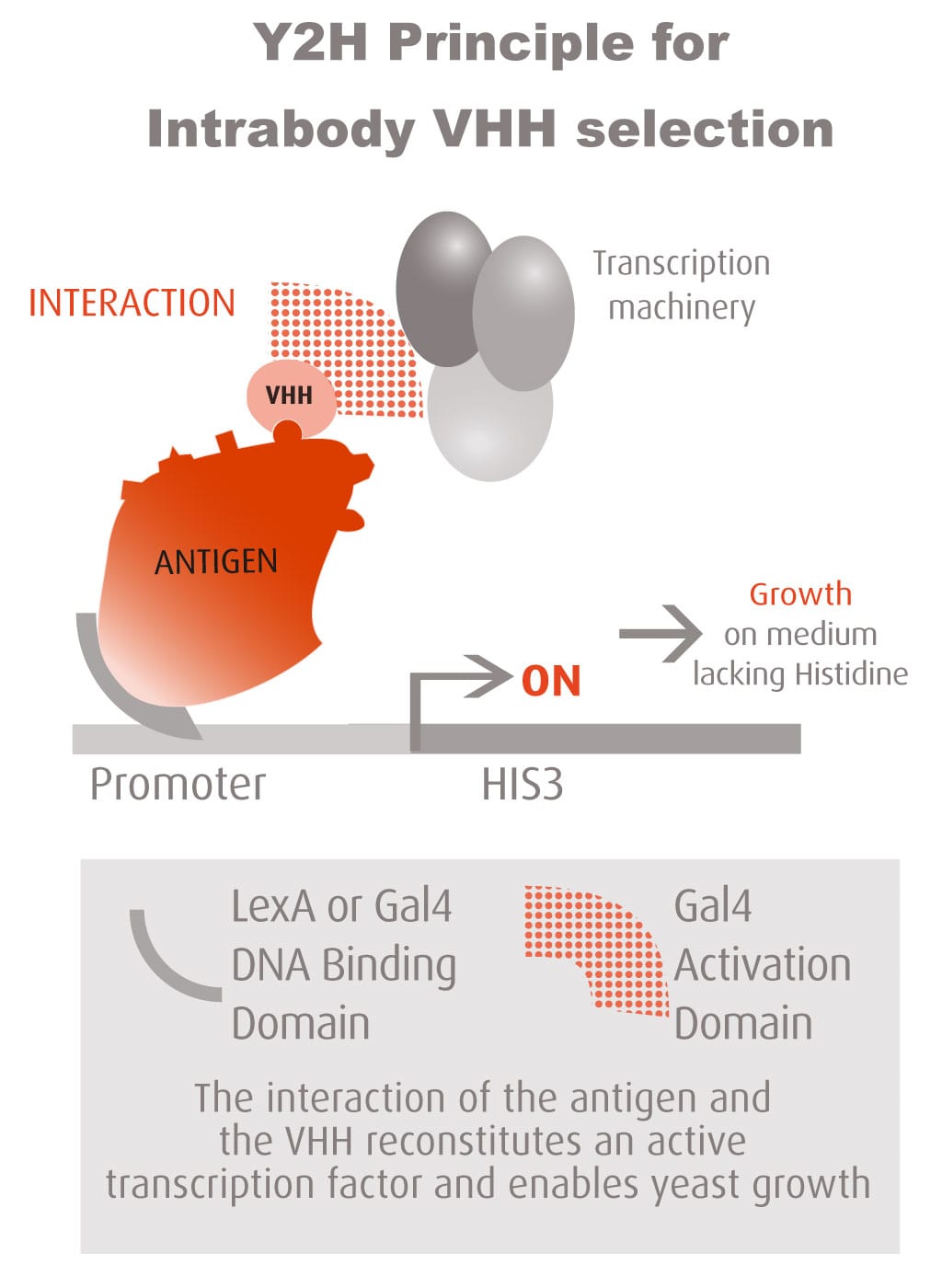
The Y2H technique is based on the reconstitution of a functional transcription factor (TF) followed by the expression of a reporter gene. This takes place in genetically modified yeast strains, in which the transcription of a reporter gene leads to a specific phenotype, usually growth on a selective medium or change in the color of the yeast colonies. Upon direct binding of the sequences of interest (bait) to protein fragments of the library (prey), the DNA Binding Domain (DBD) of the TF is brought closer to its Activation Domain (AD). The most popular fusions use the DBD and AD of the yeast TF Gal4. The bacterial protein LexA is also frequently used as a DBD in combination with Gal4 AD.
Reconstitution of the TF activates HIS3 reporter gene transcription, allowing the yeast cells to grow on a selective medium lacking for example histidine. The plasmid DNA of the positive clones is sequenced to identify the protein partners.
The development and gradual improvements of the yeast two-hybrid system (Y2H) since the early 90’s revolutionized the way protein interactions could be detected (1).
As a genetic technique, a yeast two-hybrid screen offers a sensitive and cost-effective means to test the direct interaction between two targeted proteins, or to use one’s favorite protein as a bait to screen libraries of proteins fragments prepared from the desired cell types, tissues or entire organisms. The identity of the interacting partners is then obtained by sequencing the corresponding plasmids in the selected yeast colonies. Collections of full-length proteins (‘ORFeomes’) are also becoming available for several species, but they do not cover the entire proteome yet.
The yeast two-hybrid system was rapidly adopted by the scientific community and screens in various species and research fields already led to dozens of thousands of publications. It remains the method of choice when it comes to discover novel protein interactions, as reflected by the recently published literature.
Variations of Y2H were developed to conduct screens in the presence of a co-factor, or an enzyme required for a given post-translational modification of the protein partners (2).
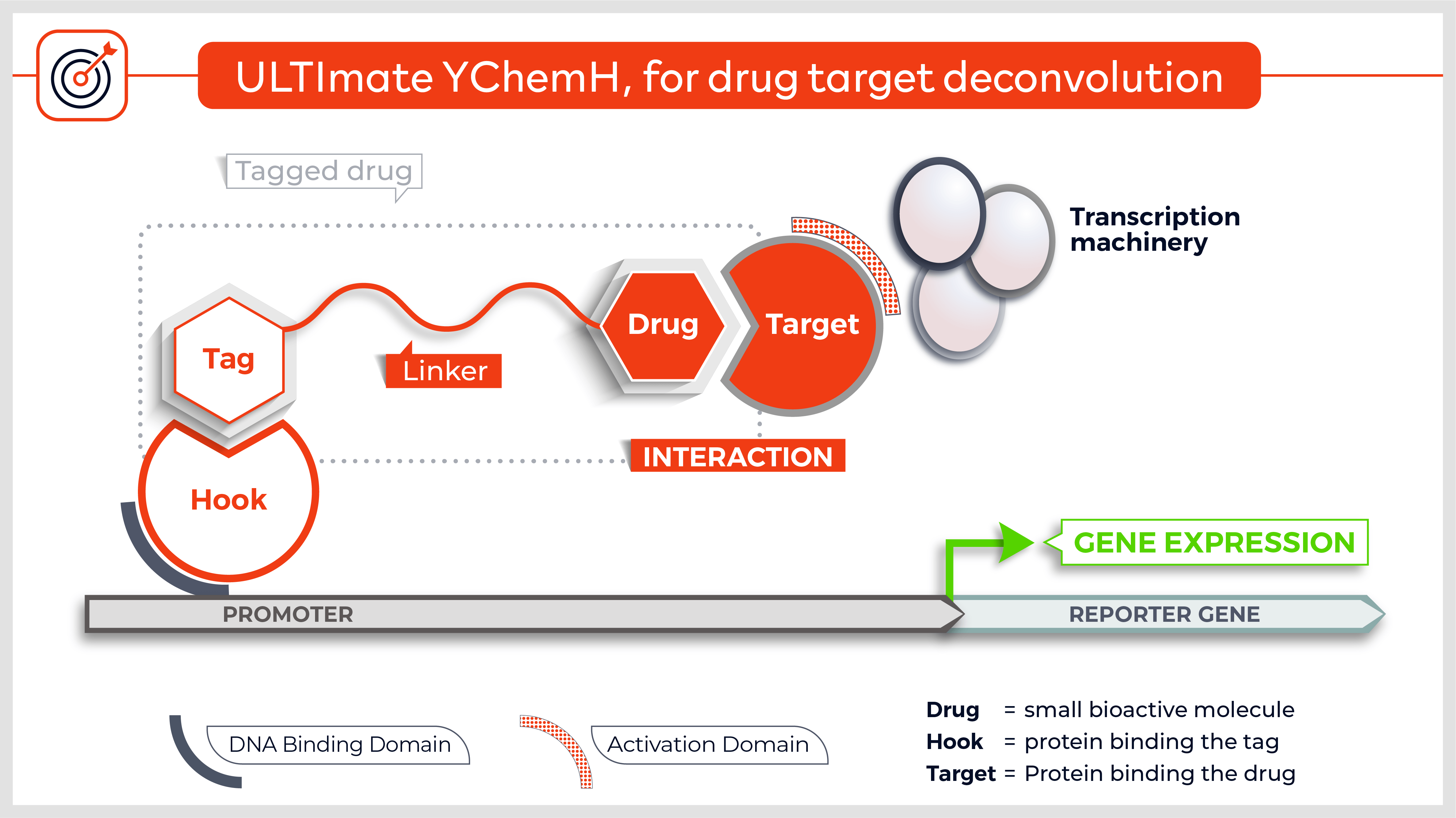
Other versions allow to screen integral membrane proteins (3) and the technique was adapted to detect protein-protein interactions in mammalian cells (4). Finally, yeast n-hybrid protocols were also devised to screen for novel DNA-protein5, RNA-protein6 and small molecule-protein interactions (7).
Contact our team of scientists for advice on your project.
References
1. Fields, S. and Song, O. A novel genetic system to detect protein-protein interactions (1989) Nature 340, 245-246
2. Naba, A., Reverdy, C., Louvard, D. and Arpin, M. Spatial recruitment and activation of the Fes kinase by ezrin promotes HGF-induced cell scattering (2008) EMBO J. 27(1), 38-50
3. Stagljar, I., Korostensky, C., Johnsson, N. and te Heesen, S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo (1998) PNAS 95(9), 5187-5192
4. Eyckerman, S. , Verhee, A., der Heyden, J.V., Lemmens, I., Ostade, X.V., Vandekerckhove, J. and Tavernier, J. Design and application of a cytokine-receptor-based interaction trap (2001) Nat Cell Biol. 3(12), 1114-1119
5. Li, J.J. and Herskowitz, I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system (1993) Science 262(5141), 1870-1874
6. Putz, U., Skehel, P. and Kuhl, D. A tri-hybrid system for the analysis and detection of RNA--protein interactions (1996) Nucleic Acids Res 24, 4838-4840
7. Licitra, E.J. and Liu, J.O. A three-hybrid system for detecting small ligand-protein
receptor interactions (1996) PNAS 93(23), 12817-12821